You are not so strong with SQL or you are not good at programming? And you need to create distributed scalable search on a very large dataset stored in HBase? And you need to achieve NRT (Near Real Time) indexing? Cloudera search along with Lily Hbase Indexer is there to rescue you!
Goal of Document
- Familiarization with Cloudera search and its components
- High level component diagram for NRT Indexing by Cloudera search
- Brief description of the work horse : Morphline
- Installation & configuration
- Basic steps of implementation for one NRT indexing example
Cloudera search and its components
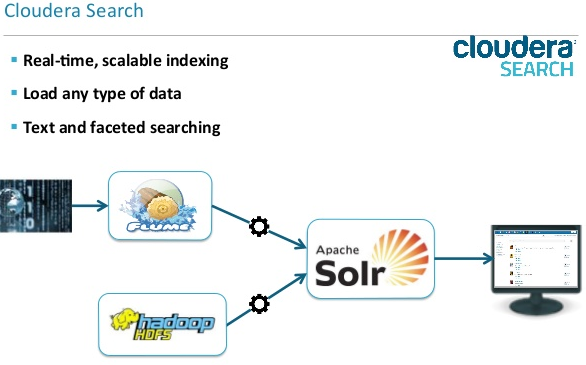
Cloudera Search is one of Cloudera's near-real-time access products. Cloudera Search enables non-technical users to search and explore data stored in or ingested into Hadoop and HBase.
Another benefit of Cloudera Search, compared to stand-alone search solutions, is the fully integrated data processing platform. Search uses the flexible, scalable, and robust storage system included with CDH. This eliminates the need to move larger data sets across infrastructures to address business tasks.
Backbone of Cloudera Search is Apache Solr, which includes Apache Lucene, SolrCloud, Apache Tika, and Solr Cell.
Few important facilities of Cloudera Search:
- Index storage in HDFS
- Provides two types of indexing ? batch and NRT
- Cloudera Search has built-in MapReduce jobs for indexing data stored in HDFS.
- Cloudera Search provides integration with Flume to support near-real-time indexing. As new events pass through a Flume hierarchy and are written to HDFS, those same events can be written directly to Cloudera Search indexers.
- NRT Indexing of HBase data using Lily HBase NRT Indexer
- The Lily HBase Indexer uses Solr to index data stored in HBase. As HBase applies inserts, updates, and deletes to HBase table cells, the indexer keeps Solr consistent with the HBase table contents, using standard HBase replication features. The indexer supports flexible custom application-specific rules to extract, transform, and load HBase data into Solr.
Now from NRT Indexing of HBase perspective:
Image courtesy: www.hadoopsphere.com
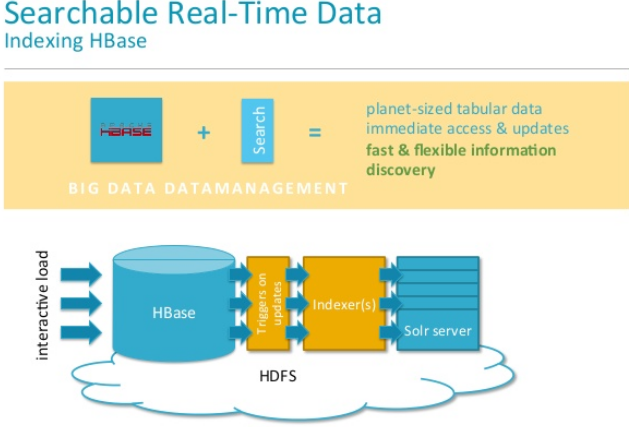
High level overview of NRT Indexing by Cloudera search and component diagram
The Lily HBase NRT Indexer Service is a flexible, scalable, fault-tolerant, transactional, near real-time (NRT) system for processing a continuous stream of HBase cell updates into live search indexes. Typically it takes seconds for data ingested into HBase to appear in search results; this duration is tunable.
The Lily HBase Indexer uses SolrCloud to index data stored in HBase. As HBase applies inserts, updates, and deletes to HBase table cells, the indexer keeps Solr consistent with the HBase table contents, using standard HBase replication.
The indexer supports flexible custom application-specific rules to extract, transform, and load HBase data into Solr.
Solr search results can contain columnFamily: qualifier links back to the data stored in HBase. This way, applications can use the Search result set to directly access matching raw HBase cells. Indexing and searching do not affect operational stability or write throughput of HBase because the indexing and searching processes are separate and asynchronous to HBase.
Image courtesy: cloudera.github.io
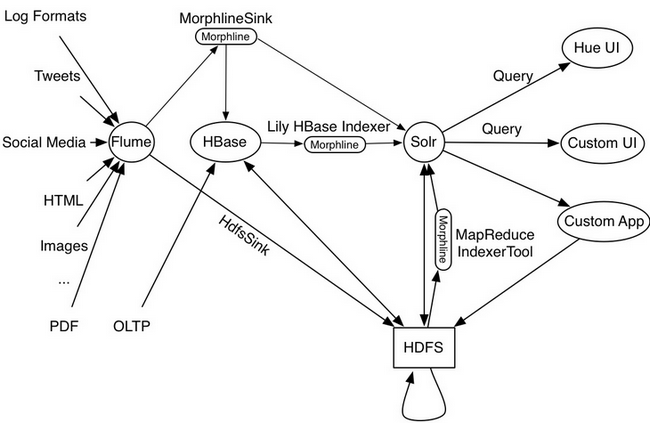
Slightly deep dive into the working of this Lily HBase Indexer reveals
Image courtesy: techkites.blogspot.in
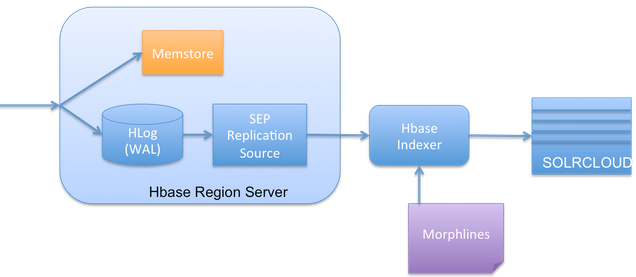
There is a well-known feature of HBase Replication where the data written into HBase is replicated to the slave HBase cluster. The Lily HBase Indexer is implemented using HBase replication, presenting (rather pretending!!) Indexers as region servers of the slave cluster.
So the indexer works by acting as an HBase replication sink. As updates are written to HBase region servers, it is written to the HLog (WAL) and HBase replication continuously polls the HLog files to get the latest changes and they are "replicated" asynchronously to the HBase Indexer processes. The indexer analyzes then incoming HBase cell updates, and it creates Solr documents (Solr documents are Solr equivalent of database row!) and pushes them to SolrCloud servers.
Hence this requires we need to make HBase replication on at the HBase cluster level (in hbase-site.xml), as well as the individual table and column family level (REPLICATION_SCOPE flag). Will show this in example implementation segment of this document.
SEP Replication Source:
SEP is Side Effect Processor. This is the listener component in HBase that listens to the changes made in HBase and enable replication asynchronously.
While most of the components are quite familiar, Morphline can be the a little lesser known but its really the work horse of the system. Will talk about it.
A brief introduction of Morphline
Cloudera Morphlines is an open-source framework that performs ETL to expedite search app development.
A morphline is a rich configuration file that simplifies defining an ETL transformation chain. Use these chains to consume any kind of data from any data source, process the data, and load the results into Cloudera Search. Executing in a small, embeddable Java runtime system, morphlines can be used for near real-time applications as well as batch processing applications. The following diagram shows the one example process flow which is defined in one morphline config file:
Image courtesy: cloudera.github.io
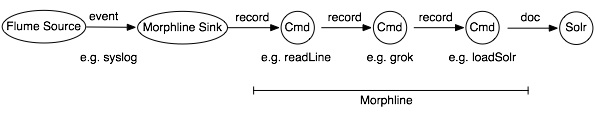
For our case, we use it to transform the HBase cell updates and map them to fields in SOLR.
The framework ships with a set of frequently used high-level transformation and I/O commands (e.g. readLine, loadSolr) that can be combined in application-specific ways. The plug-in system allows you to add new transformations and I/O commands and integrates existing functionality and third-party systems.
Scalability:
For high scalability, you can deploy many morphline instances on a cluster in many Flume agents and MapReduce tasks.
How various Cloudera components interact with Cloudera search
Please refer Cloudera Search and Other Cloudera Components
Installation & configuration using Cloudera Manager
Starting with Cloudera Enterprise 5, Cloudera Search and Lily HBase indexer will install and start by default making this process even easier. But before that you need to follow the blow steps.
- Installing the SOLR Parcel
To download the SOLR parcel directly from Cloudera, you can use the default settings for "Remote Parcel Repository URLs" (under the Parcels section in the Administration tab) as shown below
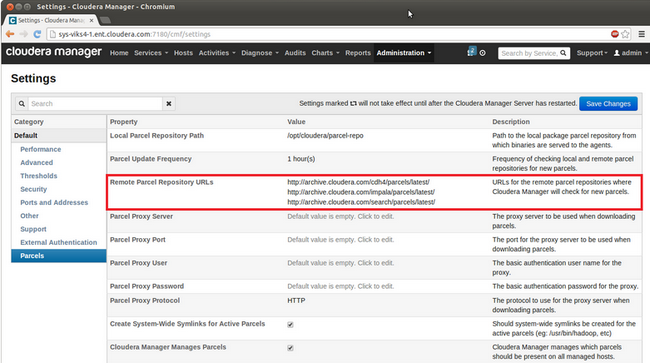
- Installing the SOLR Parcel
"Download", "Distribute," and "Activate" the parcel from the Parcels page on the Hosts tab
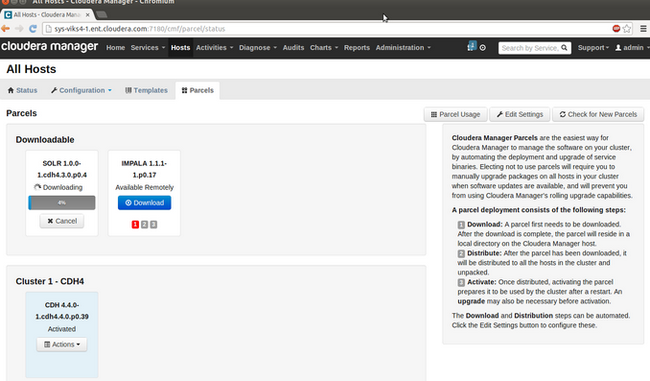
Once the parcel is activated, you have all components of Cloudera Search (Solr, Lily HBase Indexer, and Apache Flume's Morphlines Sink) ready to be used along with CDH. - Configure HBase to use Lily HBase Indexer
Before you can use the Lily HBase Indexer however, you need to ensure that replication and indexing are enabled in the HBase service in the cluster. You can change these properties on the HBase service configuration page under the "Backup" section.
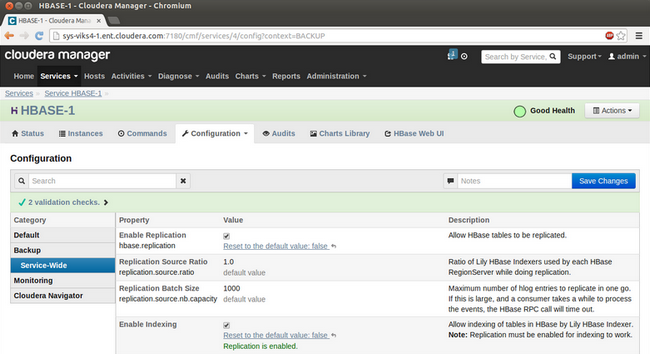
Reference: How-to: Add Cloudera Search to Your Cluster using Cloudera Manager
Installation & configuration without Cloudera manager
- Install solr-server packages using apt-get
$ sudo apt-get install solr-server - Installing and Starting ZooKeeper Server
If not done already follow Installing the ZooKeeper Packages - Initializing Solr
Configure the ZooKeeper Quorum address in /etc/default/solr. Edit the property to configure the nodes with the address of the ZooKeeper service.
ExampleSOLR_ZK_ENSEMBLE=<zkhost1>:2181,<zkhost2>:2181,<zkhost3>:2181/solr - Configuring Solr for Use with HDFS
Configure the HDFS URI for Solr to use as a backing store in /etc/default/solr. Do this on every Solr Server host:
For configuring Solr to work with HDFS High Availability (HA), you may want to configure Solr's HDFS client. You can do this by setting the HDFS configuration directory in /etc/default/solr. Do this on every Solr Server host:SOLR_HDFS_HOME=hdfs://namenodehost:8020/solrSOLR_HDFS_CONFIG=/etc/hadoop/conf - Creating the /solr Directory in HDFS
$ sudo -u hdfs hadoop fs -mkdir /solr $ sudo -u hdfs hadoop fs -chown solr /solr - Initializing ZooKeeper Namespace
$ solrctl init ?-force - Starting Solr
On Success you can see$ sudo service solr-server restart$ sudo jps -lm 31407 sun.tools.jps.Jps -lm 31236 org.apache.catalina.startup.Bootstrap start - Runtime Solr Configuration
In order to start using Solr for indexing the data, you must configure a collection holding the index. A configuration for a collection requires a solrconfig.xml file, a schema.xml and any helper files may be referenced from the xml files.
The solrconfig.xml file contains all of the Solr settings for a given collection, and the schema.xml file specifies the schema that Solr uses when indexing documents.
Configuration files for a collection are managed as part of the instance directory. To generate a skeleton of the instance directory run:
You can customize it by directly editing the solrconfig.xml and schema.xml files that have been created in $HOME/solr_configs/conf.$ solrctl instancedir --generate $HOME/solr_configs
Once you are satisfied with the configuration, you can make it available for Solr to use by issuing the following command, which uploads the content of the entire instance directory to ZooKeeper:
You can use the solrctl tool to verify that your instance directory uploaded successfully and is available to ZooKeeper. You can use the solrctl to list the contents of an instance directory as follows:$ solrctl instancedir --create collection1 $HOME/solr_configs $ solrctl collection --create collection1$ solrctl instancedir -list - Installing MapReduce Tools for use with Cloudera Search
$ sudo apt-get install solr-mapreduce - Installing the Lily HBase Indexer Service
$ sudo apt-get install hbase-solr-indexer hbase-solr-doc
Solr terminology
Before we dive into developing our small application let's get familiar with a few Solr terminologies : https://wiki.apache.org/solr/SolrTerminology
Sample Application development
Consult page: Using the Lily HBase NRT Indexer Service
Goal: As rows are added to HBase, SOLR index will be updated in NRT mode using Lily NRT HBase indexer
Steps:
- Enabling cluster-wide HBase replication
An example of settings required for configuring cluster-wide HBase replication is presented in /usr/share/doc/hbase-solr-doc*/demo/hbase-site.xml. You must add these settings to all of the hbase-site.xml configuration files on the HBase cluster, except the replication.replicationsource.implementation property which does not need to be added. - **Pointing an Lily HBase NRT Indexer Service at an HBase cluster that needs to be indexed **
This is done through Zookeeper. Add the following property to /etc/hbase-solr/conf/hbase-indexer-site.xml. Remember to replace hbase-cluster-zookeeper with the actual ensemble string as found in hbase-site.xml configuration file:<property> <name>hbase.zookeeper.quorum</name> <value>hbase-cluster-zookeeper</value> </property> <property> <name>hbaseindexer.zookeeper.connectstring</name> <value>hbase-cluster-zookeeper:2181</value> </property> - Starting an Lily HBase NRT Indexer Service
hduser@ubuntu:~$ sudo service hbase-solr-indexer restart - Start Solr
hduser@ubuntu:~$ sudo service solr-server restart - Enabling replication on HBase column families
Assuming the 'record' is our HBase table and 'data' is the only column familyhduser@ubuntu:~$ hbase shell hbase shell> create 'record', {NAME => 'data', REPLICATION_SCOPE => 1} - Creating a corresponding SolrCloud collection
This will create a folder in HOME directory containing schema file and solrconfig file among others. hbase-collection-NRT is our intended collection namehduser@ubuntu:~$ solrctl instancedir --generate $HOME/hbase-collection-NRT
Edit schema.xml to include your field (data) to be indexed.
Learn about writing schema file: http://www.metaltoad.com/blog/crafting-apache-solr-schemaxml
An entry should be something like this:<field name="data" type="text_general" indexed="true" stored="true"/>hduser@ubuntu:~$ solrctl --zk localhost:2181/solr instancedir --create hbase-collection-NRT /home/hduser/hbase-collection-NRT hduser@ubuntu:~$ solrctl --zk localhost:2181/solr collection --create hbase-collection-NRT -s 1 -c hbase-collection-NRT - Creating a Lily HBase Indexer configuration
Need to create a morphline-hbase-mapper.xml indexer configuration file which refers to refer to the MorphlineResultToSolrMapper implementation and also point to the location of a Morphline configuration file. This config file holds the morphline commands and the mapping between HBase column and solr-index
Note that this also contains the reference to the Hbase table ('record' in our case) to be indexedhduser@ubuntu:~$ cat $HOME/morphline-hbase-mapper.xml <?xml version="1.0"?> <indexer table="record" mapper="com.ngdata.hbaseindexer.morphline.MorphlineResultToSolrMapper"> <!-- The relative or absolute path on the local file system to the morphline configuration file. --> <! -- Use relative path "morphlines.conf" for morphlines managed by Cloudera Manager --> <param name="morphlineFile" value="/etc/hbase-solr/conf/morphlines.conf"/> <!-- The optional morphlineId identifies a morphline if there are multiple morphlines in morphlines.conf --> <! -- <param name="morphlineId" value="morphline1"/> --> </indexer> - Creating a Morphline Configuration File
Now we have to implement the indexer config file that is referred in previous step. At minimum, there are three important things you must notice:- This file contains actual morphline commands that perform the ETL work. In our case the main command is extractHBaseCells
- Mapping between the Hbase column and corresponding target Solr schema field
- Morphline id; this will be required if we decide to write multiple morphline flow in same file. In our case we have only one flow; still we referred to that flow using id (morphline1).
hduser@ubuntu:~$ cat /etc/hbase-solr/conf/morphlines.conf morphlines : [ { id : morphline1 importCommands : ["org.kitesdk.morphline.**", "com.ngdata.**"] commands : [ { extractHBaseCells { mappings : [ { inputColumn : "data:*" outputField : "data" type : string source : value } ] } } { logTrace { format : "output record: {}", args : ["@{}"] } } ] } ]
- Registering a Lily HBase Indexer configuration with the Lily HBase Indexer Service
Here we are to create an indexer using the conf file (morphline-hbase-mapper.xml) created in step f. Note that we also need to specify the zookeeper instance and the target solr collection where indexer will load the index to.
We have used newlines for better understanding only; in command prompt use this as a single line command.hduser@ubuntu:~$ hbase-indexer add-indexer --name myNRTIndexer --indexer-conf $HOME/morphline-hbase-mapper.xml --connection-param solr.zk=localhost:2181/solr --connection-param solr.collection=hbase-collection-NRT --zookeeper localhost:2181
On successful execution we see in command centerIndexer added: myNRTIndexer - Creating a corresponding SolrCloud collection
Note:hduser@ubuntu:~$ hbase-indexer list-indexers SLF4J: Class path contains multiple SLF4J bindings. SLF4J: Found binding in [jar:file:/usr/lib/hbase-solr/lib/slf4j-log4j12-1.6.1.jar!/org/slf4j/impl/StaticLoggerBinder.class] SLF4J: Found binding in [jar:file:/usr/lib/zookeeper/lib/slf4j-log4j12-1.6.1.jar!/org/slf4j/impl/StaticLoggerBinder.class] SLF4J: See http://www.slf4j.org/codes.html#multiple_bindings for an explanation. ZooKeeper connection string not specified, using default: localhost:2181 Number of indexes: 1 myNRTIndexer + Lifecycle state: ACTIVE + Incremental indexing state: SUBSCRIBE_AND_CONSUME + Batch indexing state: INACTIVE + SEP subscription ID: Indexer_myNRTIndexer + SEP subscription timestamp: 2015-05-09T21:11:15.629+05:30 + Connection type: solr + Connection params: + solr.collection = hbase-collection-NRT + solr.zk = localhost:2181/solr + Indexer config: 576 bytes, use -dump to see content + Batch index config: (none) + Default batch index config: (none) + Processes + 1 running processes + 0 failed processes- For the newly created indexer process to running (yellowed section above) we must start the hbase-solr-indexer service as told in step c.
- Morphline configuration files can be changed without recreating the indexer itself. In such a case, you must restart the Lily HBase Indexer service.
- Verifying the indexing is working
Add rows to the indexed HBase table. For example:
If the put operation succeeds, wait a few seconds, then navigate to the SolrCloud's UI query page (http://localhost:8983/solr), and query the data.hduser@ubuntu:~$ hbase shell hbase(main):001:0> put 'record', 'row5', 'data', 'value5' hbase(main):002:0> put 'record', 'row6', 'data', 'value6'
We expect that you would see something akin to below- Core overview (you must select appropriate core from lhs dropdown)
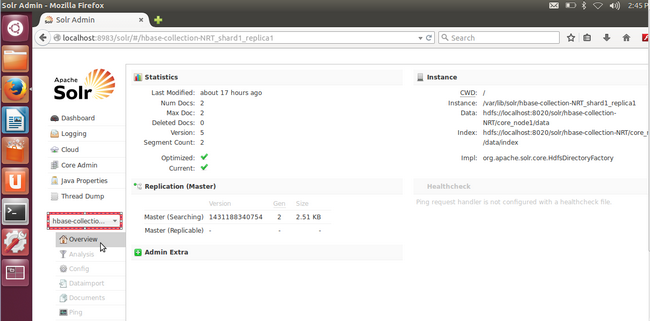
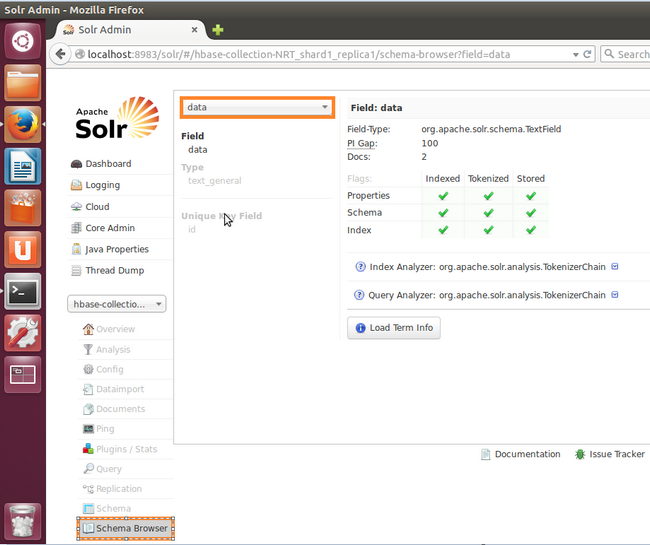
- Check that the only field (name was 'data') that we added to our schema file is featured in Schema browser. That will ensure that the correct collection is currently active in Solr. (remember to select the field name 'data' from the top dropdown)
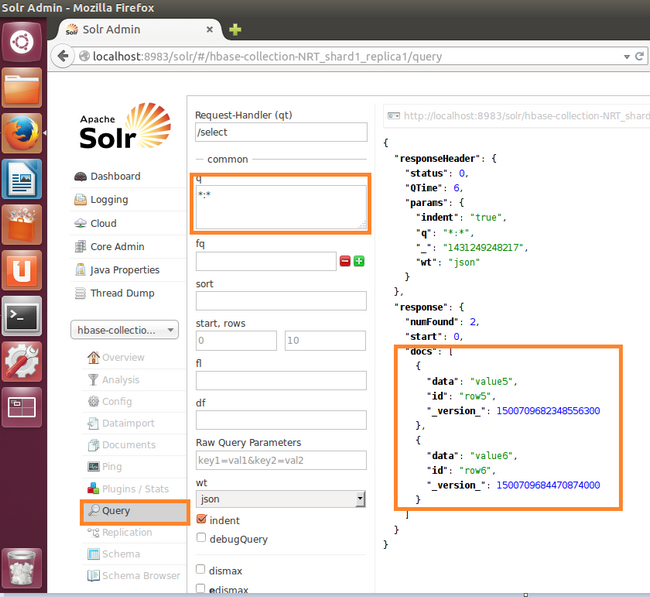
- Finally, the query on the index revels that we have two indexed rows!!!
- Core overview (you must select appropriate core from lhs dropdown)
A troubleshooting
While creating and uploading the solr collection into zookeeper I kept on getting the below error:
org.apache.solr.client.solrj.impl.HttpSolrServer$RemoteSolrException:Error CREATEing SolrCore 'hbase-collection-NRT_shard1_replica1': Unable to create core: hbase-collection-NRT_shard1_replica1 Caused by: Specified config does not exist in ZooKeeper:hbase-collection-NRT
Solution:
- Stop Solr cluster
hduser@ubuntu:~$ sudo service solr-server stop - Need to make SOLR_ZK_ENSEMBLE property value correct in /etc/default/solr file. For my case it should be
SOLR_ZK_ENSEMBLE=localhost:2181/solr - Reinitialize the solr cluster, That would clear the Solr data in ZooKeeper and hence all the collections will be removed
Then you need to restart Solr cluster and recreate the collection as described in step f of Sample Application development section.hduser@ubuntu:~$ solrctl init --force













Comments